Introducing
VarSy – The Variant Synonymizer
What is VarSy?
VarSy resolves ambiguous variant notation into a canonical identity with synonyms. The resolved identity and synonyms are then used by Variant Teller to aggregate evidence. It produces a real-time, human-readable summary. It compiles domain annotations (PANTHER, PFAM), 3D structure position, allele frequencies, and mutation or splice predictors. It compares transcript positions, notes protein nomenclature differences, and indexes genetic and functional literature via Adenine AI. It also highlights LOF evidence relevant to the gene.
Why Use VarSy?
- Unify variant names from different databases and publications.
- Quickly retrieve all synonyms for a given change.
- Streamline both clinical and research variant lookups.
- One click: Variant Teller™ aggregates evidence and delivers a real-time summary.
Query And Results
Upon successful login, query in the search bar (see the snapshot below) to retrieve exact or possible matches.
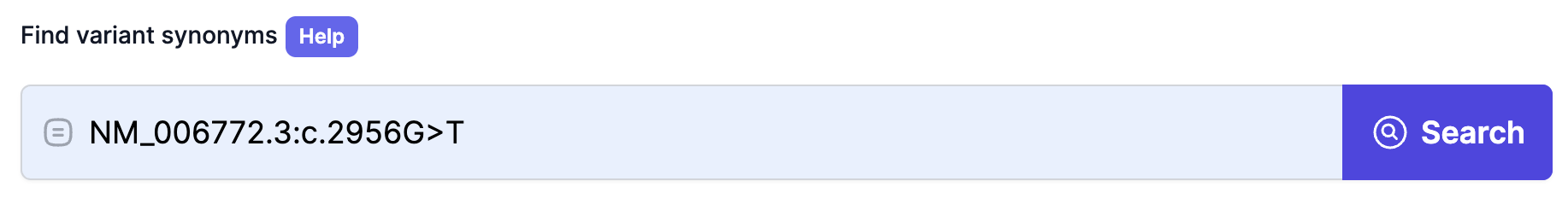
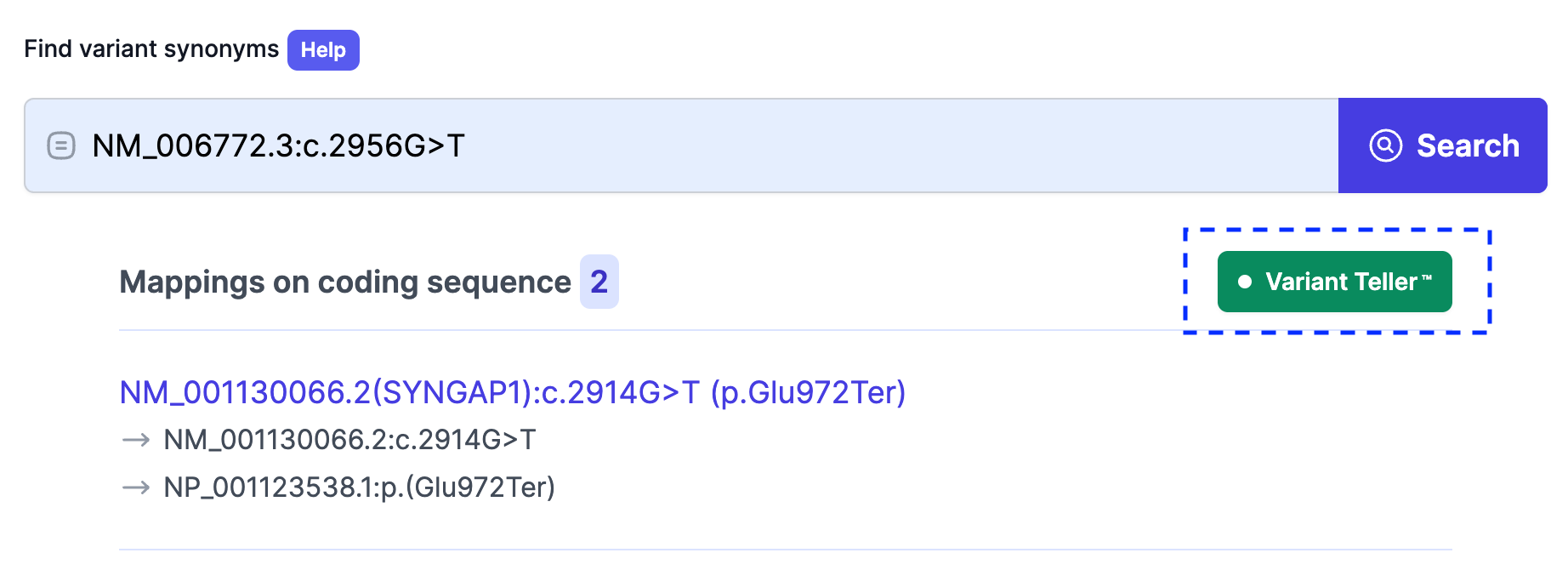
Below is an example of an exact HGVS search—here we queried “NM_006772.3:c.2956G>T” and retrieved all matching variant synonyms with their genomic, transcript, and protein mappings.
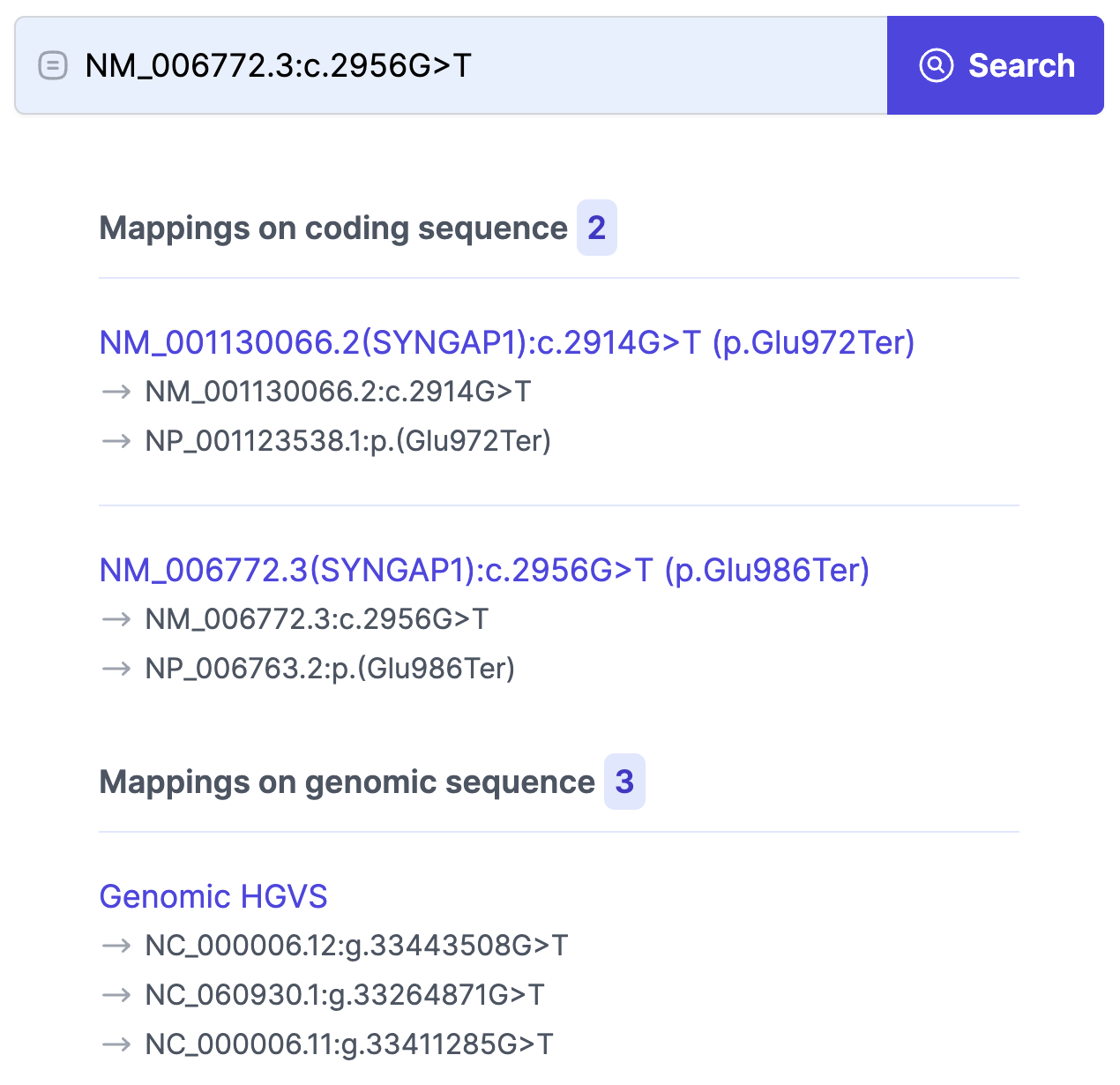
Here’s an example of an approximate query—entering “GRIN2A:Asp192Asn” returns all related variants across transcripts and nomenclatures for the GRIN2A gene.
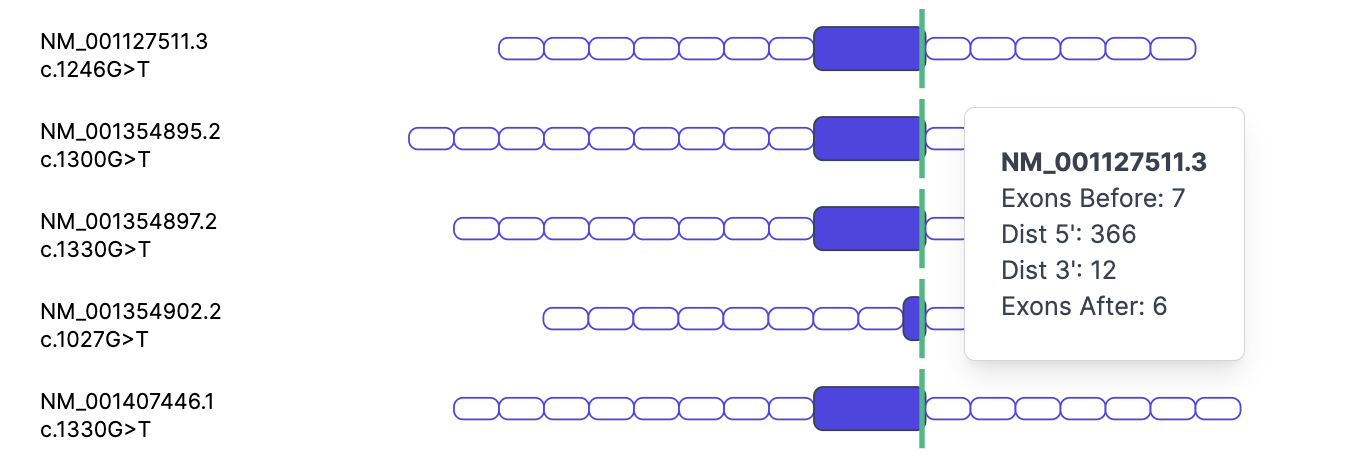
This graphical overview, included with each mapping, lets you easily spot variants that may warrant further investigation. It displays variant positions across transcript isoforms, shows exon sizes, and indicates the number of exons before and after each variant’s exon, as well as the number of bases upstream and downstream of the variant within that exon.
Examples
| Search Term | Description |
|---|---|
NM_000487.6:c.343G>T |
Transcript-level coding change on NM_000487.6 |
NP_000478.3:p.Ala115Ser |
Protein-level substitution on NP_000478.3 |
NC_000022.11:g.50627288C>A |
Genomic change on GRCh38 chr22 |
ARSA:c.343G>T |
Gene-level coding prefix for ARSA |
GRIN2A:p.Asp192Asn |
Protein-level substitutions for GRIN2A |
Variant Teller
Once VarSy resolves the variant’s identity and synonyms, click Variant Teller™. It aggregates evidence and delivers a real-time, transcript-aware summary. Includes domains (PANTHER, PFAM), 3D position, allele frequency, predictors, Adenine AI literature, and LOF evidence.
Summary
In summary, VarSy streamlines variant discovery by unifying disparate nomenclatures into a single, user-friendly interface. Whether you’re a clinician validating a patient’s mutation or a researcher exploring gene‐level variation, VarSy’s powerful search capabilities and clear visualizations let you access all relevant synonym mappings in seconds. Explore the UI, tap into our RESTful APIs for programmatic integration, and see how VarSy can simplify your genetic variant workflows starting today.
Variant Teller™ turns VarSy’s resolved identities into a real-time, human-readable summary. One click aggregates domains (PANTHER, PFAM), 3D position, and allele frequencies. It includes mutation and splice predictors, transcript comparisons, and protein nomenclature differences. Adenine AI supplies indexed genetic and functional literature, plus LOF-related evidence. The result is faster reviews and clearer, defensible variant decisions.